A Glossary of Parallel Computing Packages in R
There are many packages in R that facilitate parallel computing. In this post,
I intend to summarize some of the most popular functions/packages for this
purpose.
1. The built-in parallel package
The package parallel is included in R.
1.1 mclapply
If you are using Linux/Unix machines, mclapply might be your best friend for
parallelizing simple jobs on your local machine. A precise word to describe
mclapply is given by the authors in the package manual - “ephemeral”. The
basic language is like the following:
result <- mclapply(1:n,function = function(x){
...
}, mc.cores = k)
k is the number of cores you want to use simultaneously. It is ephemeral in
the sense that, after all replicates are performed, the temporary cluster of k
cores is destructed.
Note: according to the package documentation, mclapply relies on forking,
which makes it not applicable on Windows machines.
1.2 clusterApply
Compared to mclapply, clusterApply is not that ephemeral. When calling
makeCluster, you are building a cluster of 10 cores. After setting up the
cluster, the variables and functions in the workspace need to be exported to
each node, therefore clusterExport needs to be called. The grammar is as
follows:
cl <- makeCluster(k) # make a cluster of k cores
clusterExport(cl, varlist = ls()) # send variables to each node
Result <- clusterApply(cl, 1:n, fun = function(x) …)
There is a package called ParallelLogger that is built to provide more
enhanced functionals to the clusterApply function. Specifically,
ParallelLogger::clusterApply provides a progress bar which lets you monitor
your simulation progress. To me it is very important when I test my code on my
laptop. If you are using a big cluster like an HPC, however, this might not be a
big deal.
One important thing to keep in mind is to stop the cluster after the job is done, or memory leaks can happen! To stop a cluster and free up resource, call
stopCluster(cl)
Or, if the cluster construction is inside your code, you can use
on.exit(stopCluster(cl)) to ensure that the cluster is stopped upon finishing.
2. furrr
The superpower of furrr is maximized when you are used to the tidy verse
language. While map and its variants in the purrr package provides the
tidyverse version of lapply, future_map and its variants, in furrr,
provides the tidyverse counterparts of mclapply.
The “verse” is like this:
library(furrr)
plan(multiprocess(workers = k)) # number of cores
result <- future_map(1:n, .f = function(x){
...
})
Or, the ~ operator in purrr can also be used. For example (maybe a bad
example), with a list of vectors, suppose we want to find the medians of each
vector, we can do:
result <- future_map(dataList, ~ median(.x))
This might be a bad example as finding the median can be done quite quickly without using any parallelization. However, when a single job is too big, it is definitely worth it.
3. foreach
The foreach package is provided by Microsoft R, previously known as Revolution
R. The basic grammar of execution in foreach is as follows:
x <- foreach(i = 1:3) %do% sqrt(i)
To parallelize, we replace %do% with %dopar%:
registerDoParallel(k)
x <- foreach(i = 1:1000) %dopar% sqrt(i)
4. A small comparison
I intend to provide a small comparison of computing speed on my own laptop, which has Intel(R) Core(TM) i7-8850H CPU @ 2.60GHz, with 12 logical cores.
First, a list of containing 100 survival datasets, each of size 200000, is
generated using the genData function from
here.
Then for each dataset, a Cox regression is fitted.
library(microbenchmark)
library(survival)
library(furrr)
library(parallel)
library(foreach)
library(doParallel)
dataList <- purrr::map(1:100, ~updateCoxSurv::genData(0.02,
c(0.7, -0.5, 0.4),
200000, 60, 0.1))
plan(multiprocess(workers = 10))
a <- microbenchmark(future_map(dataList, .f = function(x){
coxph(Surv(survtime, status) ~ ., data = x)
}), times = 10)
b <- microbenchmark(mclapply(dataList, function(x){
coxph(Surv(survtime, status) ~ ., data = x)
}, mc.cores = 10), times = 10)
registerDoParallel(10)
c <- microbenchmark(foreach(i=1:100) %dopar% coxph(Surv(survtime, status) ~ .,
data = dataList[[i]]),
times = 10)
As can be seen each operation is repeated 10 times. Here’s a boxplot of the results.
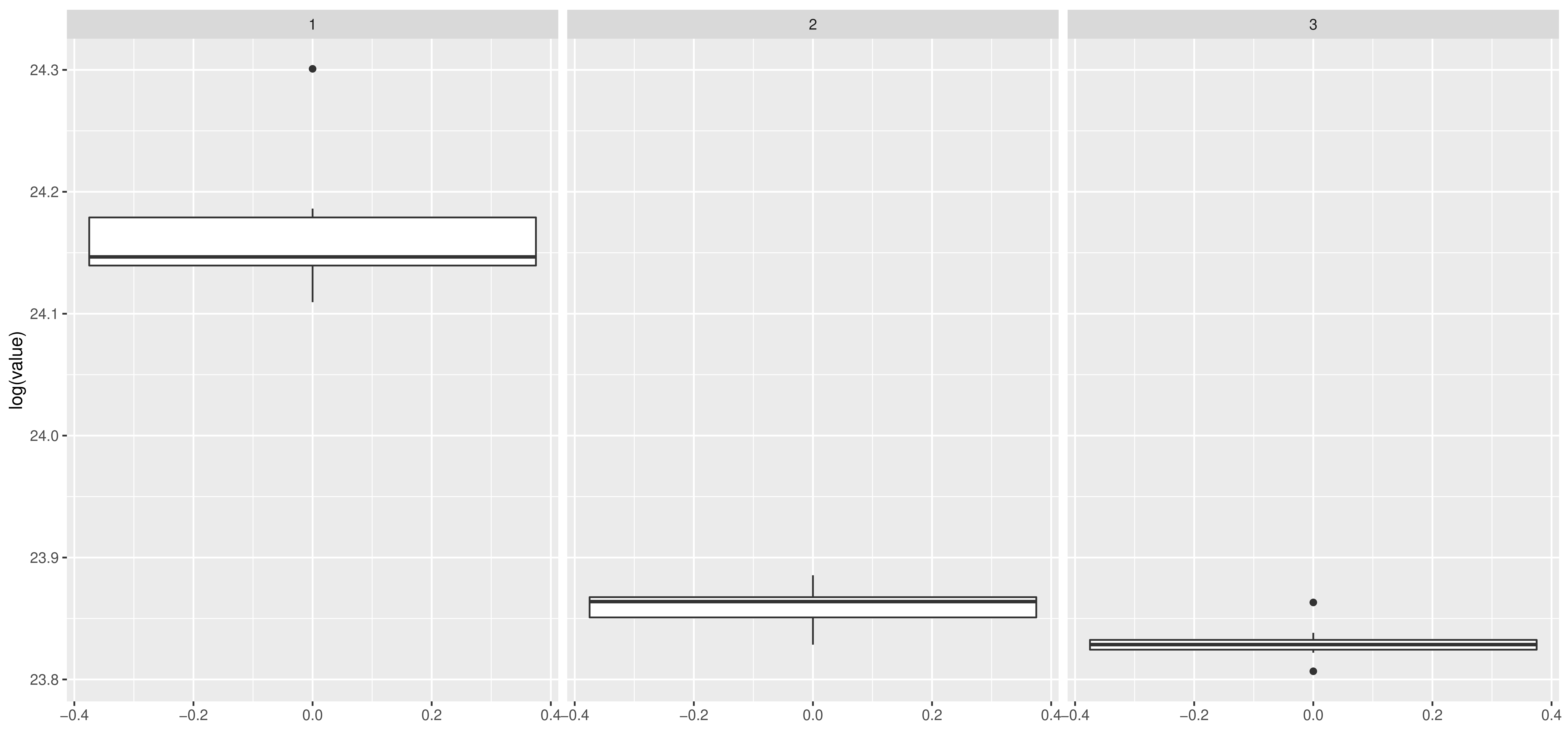
The y-axis is the logarithm of computing time in nanoseconds (10-9).
%dopar% turns out to be the fastest one, followed by mclapply and future_map. The difference, however, is very minimal.